Selecting a Workflow

Choose a Workflow
- Placement workflow (Fast): Automatically places uploaded genomes into a pruned GTDB-based reference tree using Mash distance. Suitable for quick, high-level placements.
- De novo workflow (Accurate): Allows custom tree construction using selected organisms and genes. Suitable for in-depth phylogenetic analysis.
Placement (Lite) Workflow
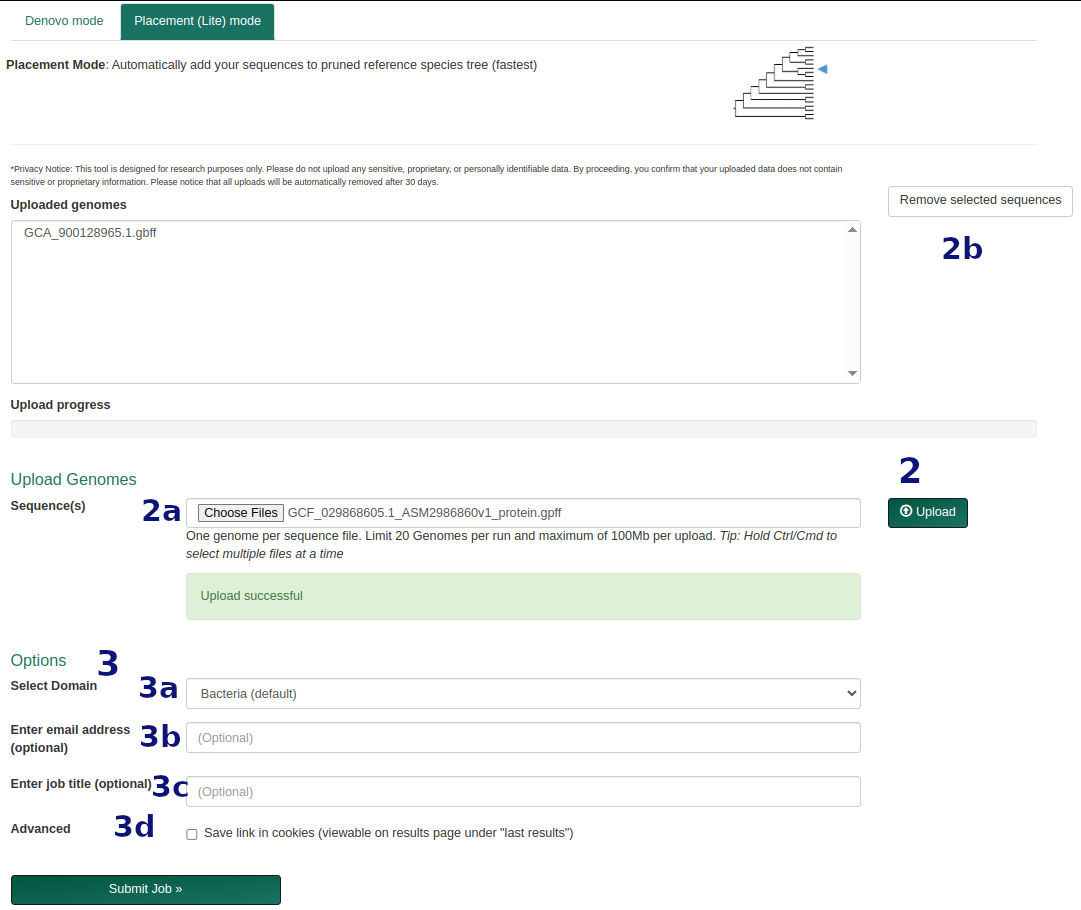
- Upload genome file(s) in FASTA or GenBank each containing one organism.
- Optional: provide an email for job updates, a title, or save the job to your browser (via cookies).
De novo Workflow
Step 1 Submit Sequences
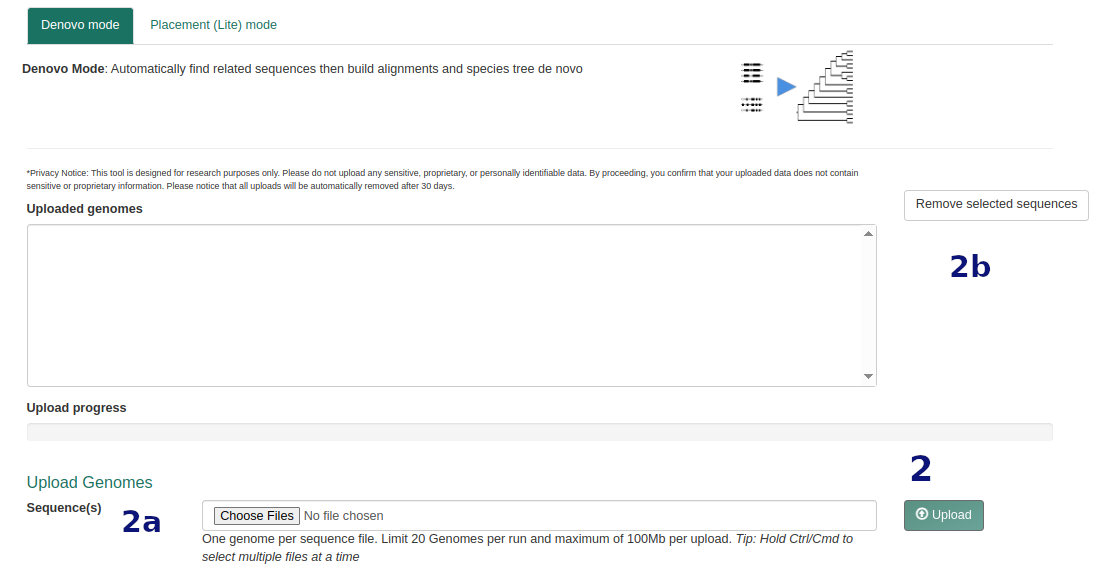
- Upload up to 50 genome files. Same format requirements as above.
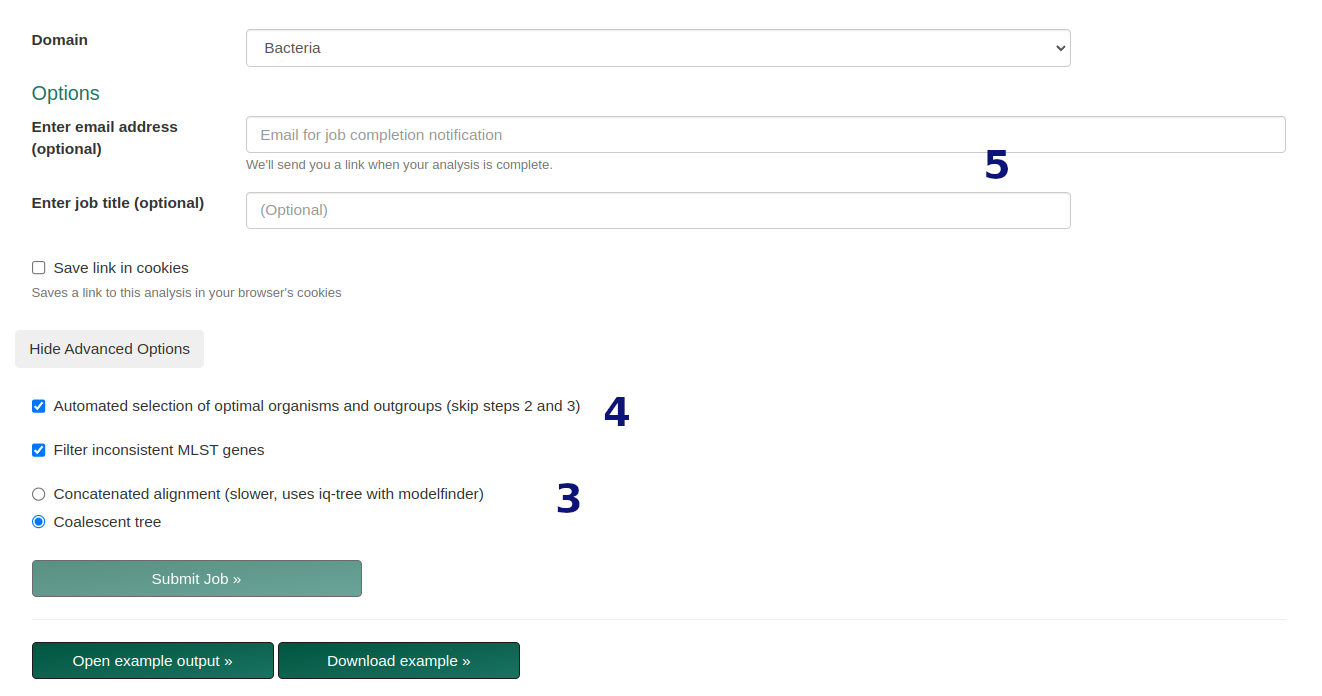
- Choose tree type:
- Concatenated: Concatenated Phylogeny, using concatenated matrix.
- Coalescent: Default MLST tree, based on gene tree consensus.
- Optional parameters:
- Enable automatic species/gene selection.
- Enable bootstrapping (Ultrafast Bootstrap).
- Enable ModelFinder for best-fit models.
- Optional job parameters (email, title, cookie storage).
Step 2 Species and Outgroup Selection
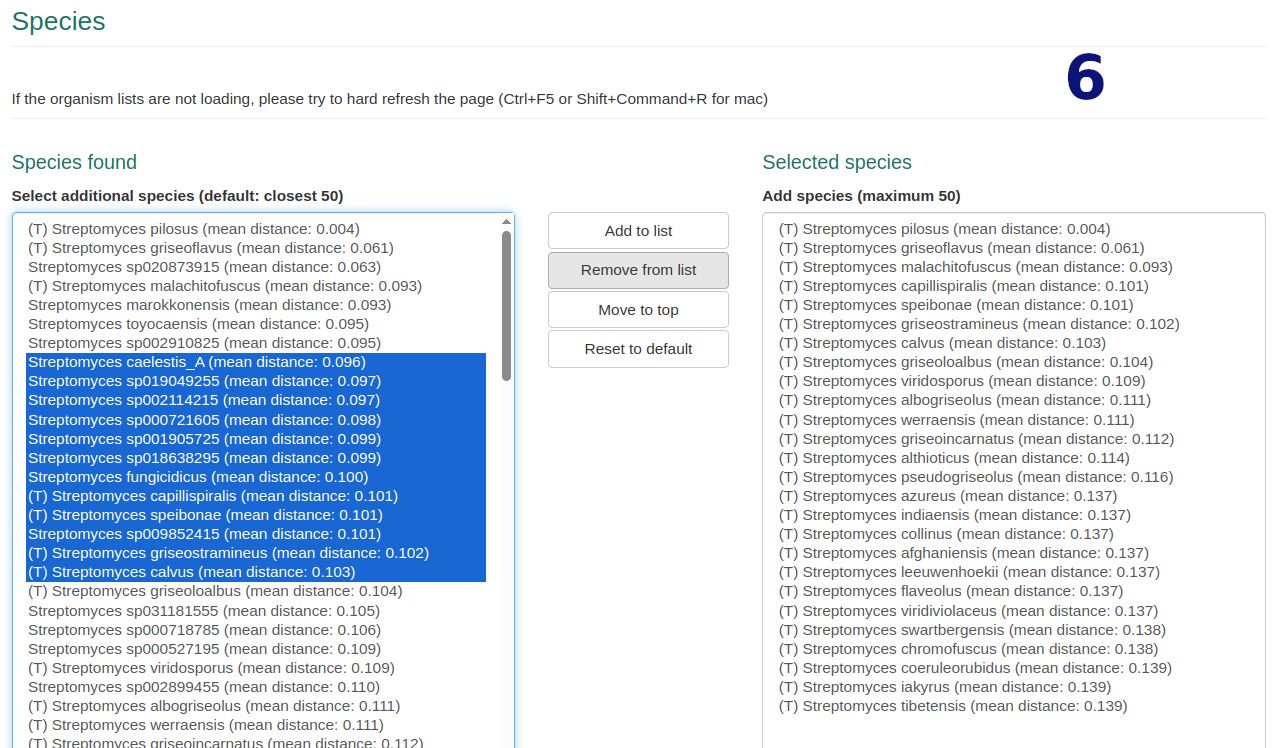
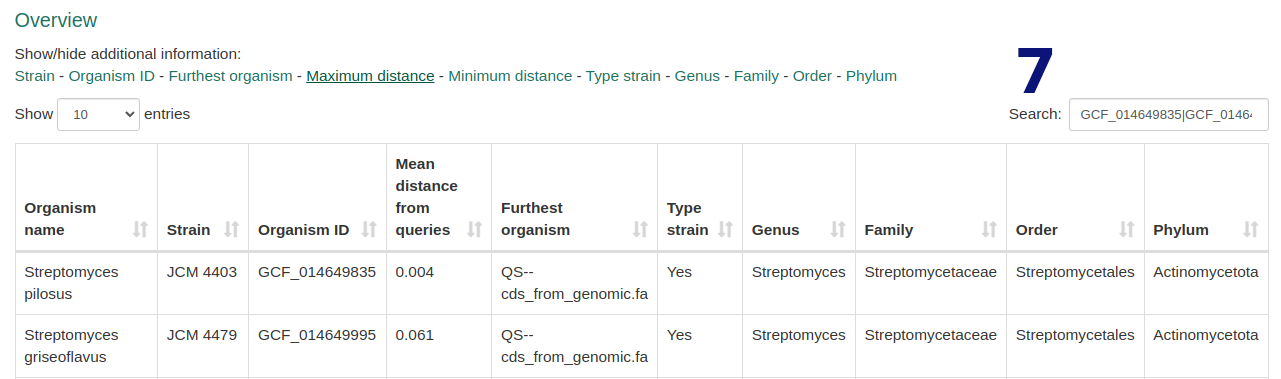
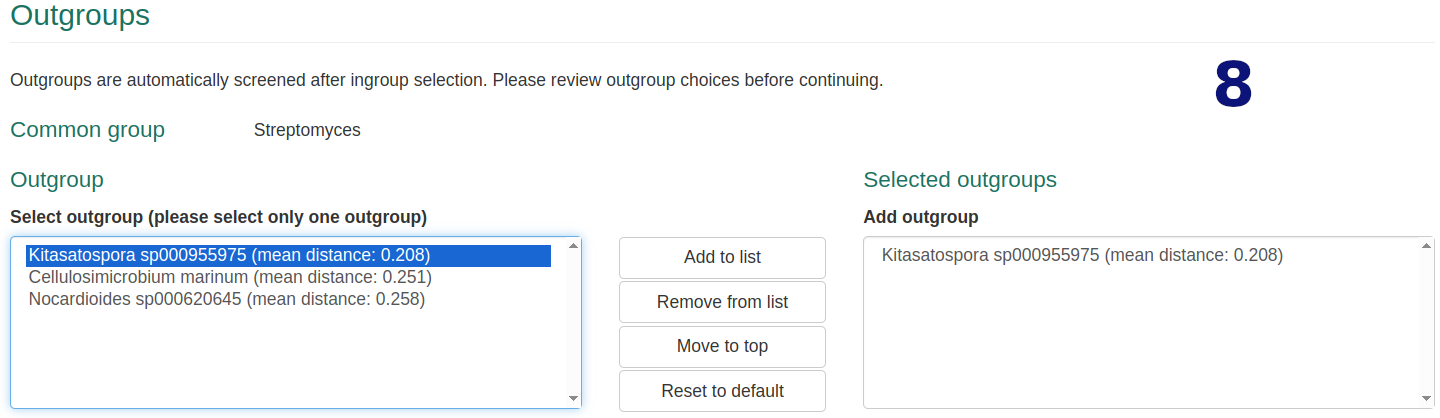
- Edit default selection of 50 closest genomes. Exceeding selections are ignored.
- Filter, search, and explore metadata for selected and additional species.
- Optionally select your desired outgroup genome.
Results
Mash-based ANI Estimation
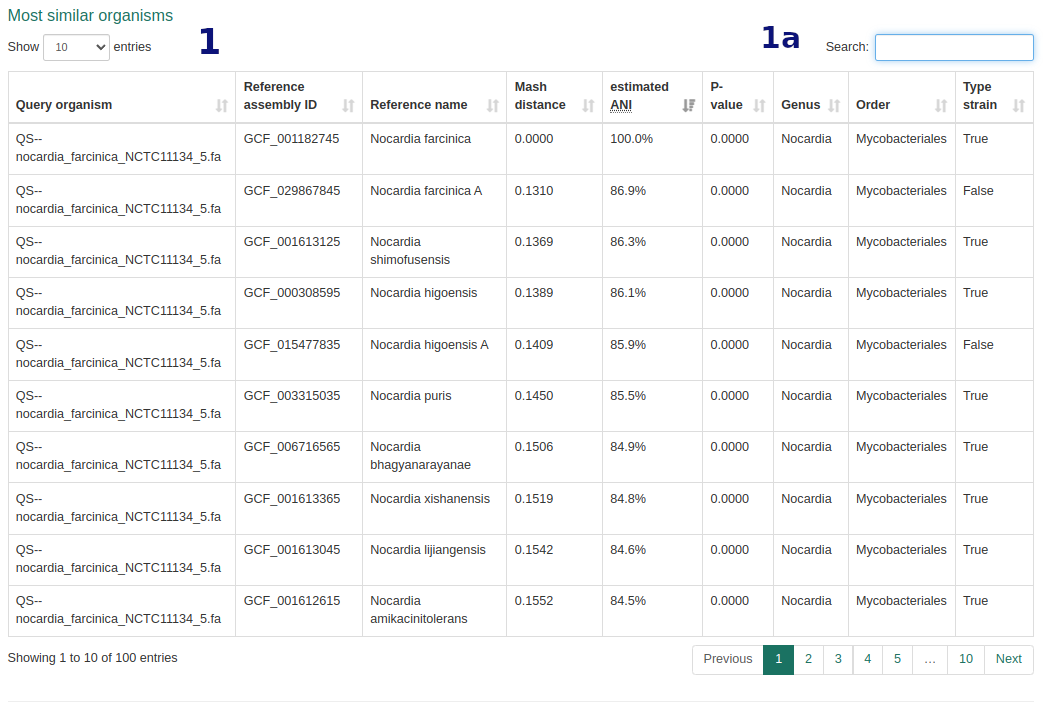
- ANI table shows closest matches by Mash distance. Search and sort functions available.
Tree
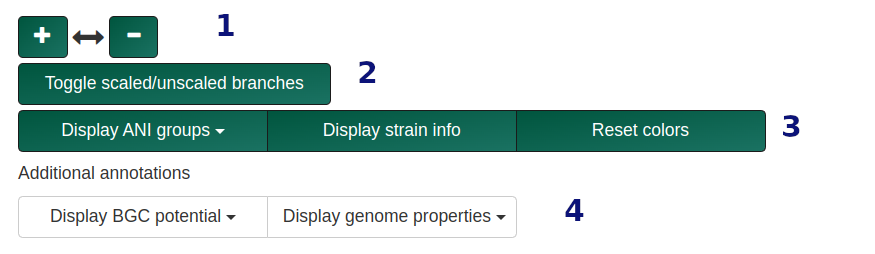
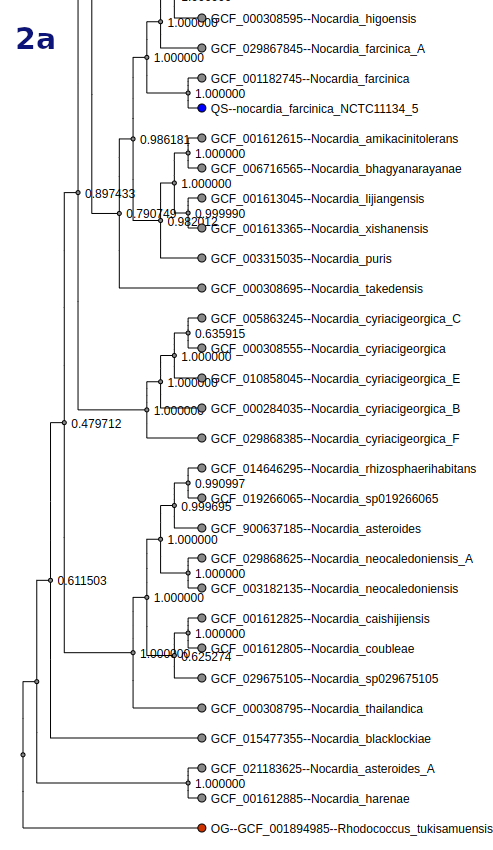
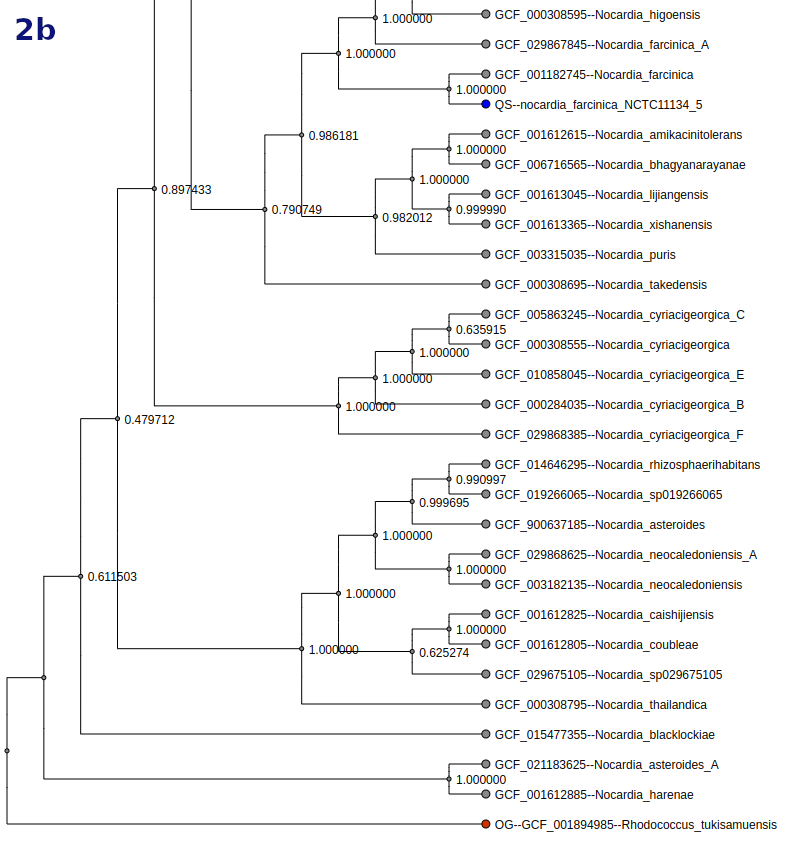
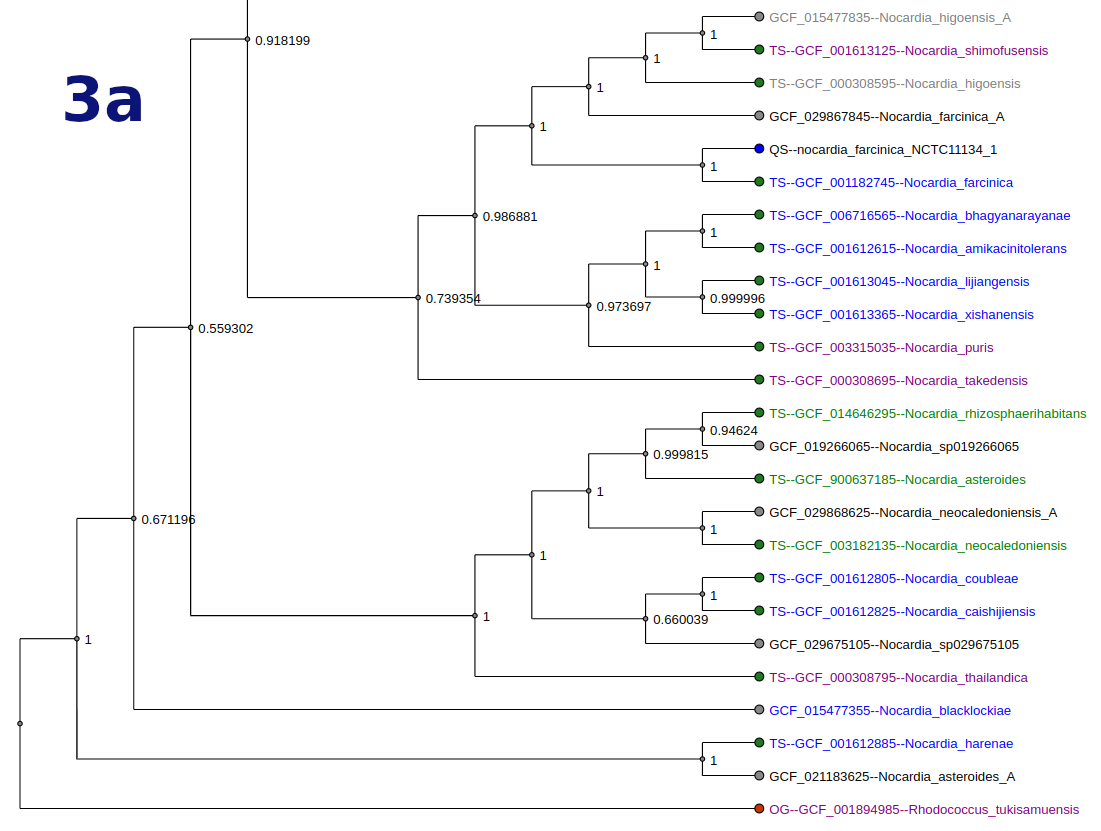
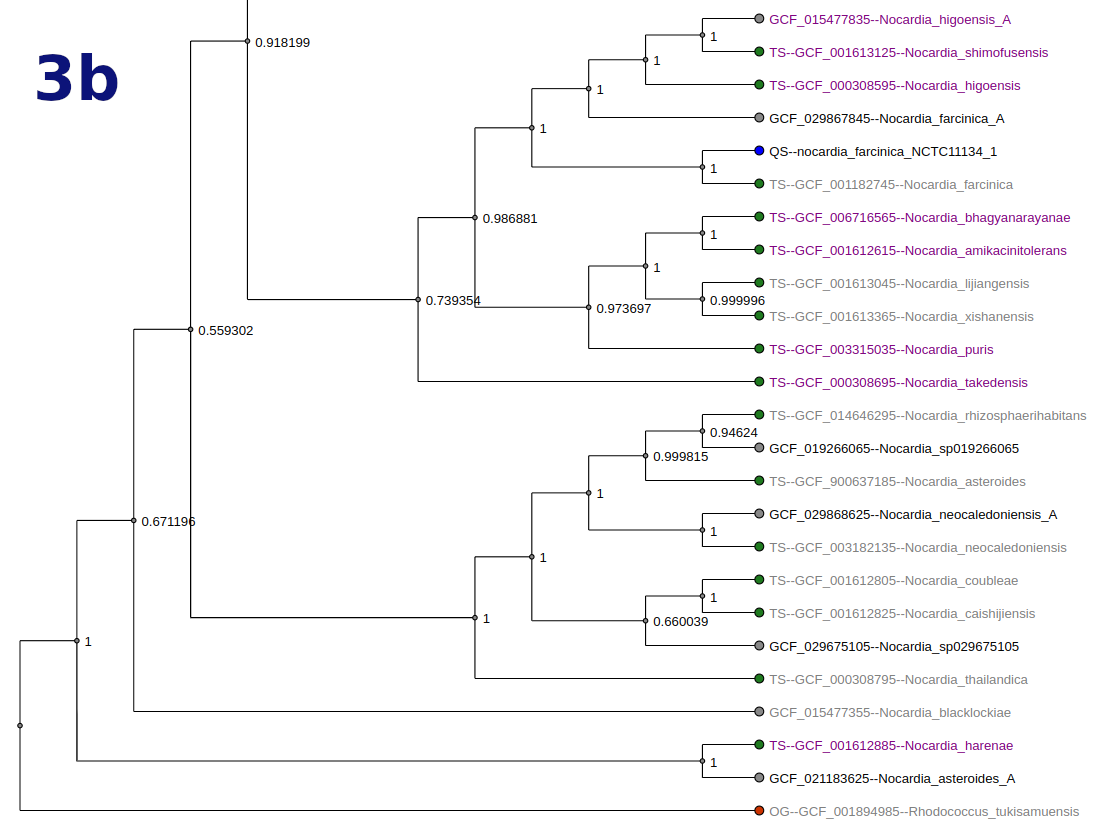

Tree types and options vary by workflow. Tree nodes are color-coded (QS = blue, OG = red, TS = green).
- Toggle between scaled and unscaled branch display.
- Search and highlight organisms in the tree.
- Display options include:
- Highlighting BGC potential
- Strain info highlighting
- Highlighting a specific BGC group
- Additional data about reference genomes
- Downloads:
- Newick tree (no color)
- SVG tree image
- Alignment files (ZIP) (denovo only)
- Gene list (denovo only)
- ANI table (full)
Related Publication
AutoMLST2 is described in our recent publication in Nucleic Acids Research:
Pourmohsenin et al. (2025). AutoMLST2: a web server for phylogeny and microbial taxonomy . Nucleic Acids Research 53(W1):W45–W54.
Cookies
We use browser cookies to optionally store links to recent jobs. These are only used to help you return to your results and are not shared or tracked.