Overview
Rapid ANI estimation

Nearest reference organisms (GTDB) are identified and collated.
MLST gene selection
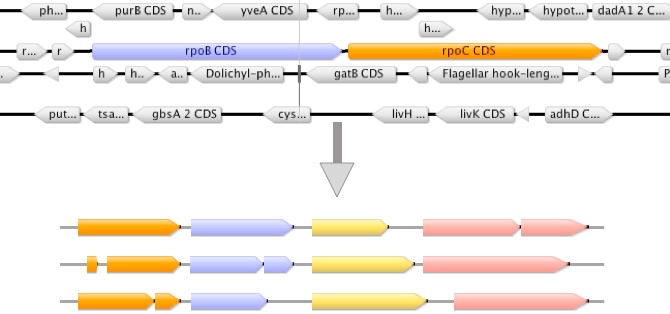
Single copy genes are identified and prioritized.
Tree building
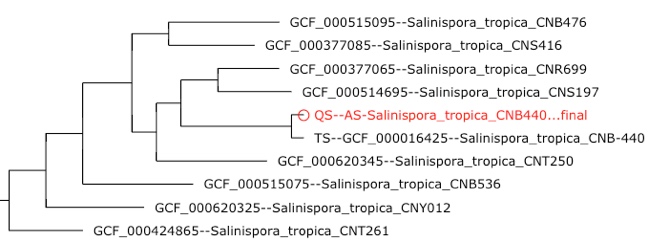
Phylogeny is built using all MLST genes from query and reference organisms using either a concatenated gene or coalescent tree method.